Key Points
- Perturb-FISH enhances gene circuitry analysis by preserving spatial relationships between cells.
- Developed at the Broad Institute, it combines CRISPR screening with spatial transcriptomics.
- The method revealed the effects of autism-related genes on brain cells and tumor-immune interactions.
- The technology can analyze 35 genes but aims to expand its capabilities further.
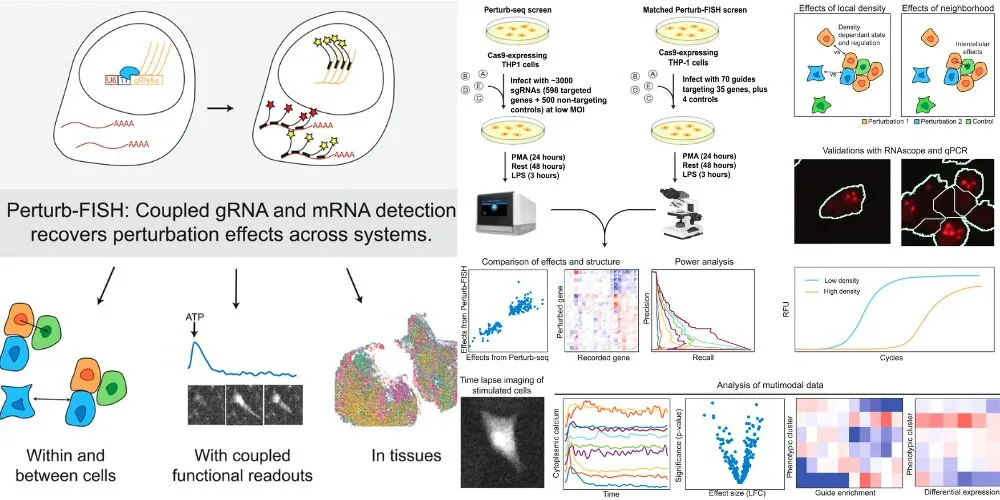
Scientists have developed a cutting-edge technology called Perturb-FISH that enhances the ability to study gene circuitry within individual cells while preserving spatial context. This innovation, created at the Spatial Technology Platform at the Broad Institute of MIT and Harvard, builds on previous gene suppression techniques by incorporating imaging-based spatial transcriptomics and large-scale CRISPR guide RNA detection.
Traditional gene editing methods, such as Perturb-seq, allow researchers to suppress and measure their effects on other genes. However, these techniques fail to capture the spatial relationships between cells, which is crucial for understanding how genes function in health and disease. Perturb-FISH addresses this limitation by enabling scientists to track how genetic changes in one cell influence neighboring cells.
The research was published in Cell. Using this method, researchers investigated the impact of autism-related genes on human brain cells known as astrocytes, revealing changes in calcium activity and gene expression. Additionally, they studied tumor-immune cell interactions in an animal model, demonstrating the tool’s effectiveness in complex biological systems.
Perturb-FISH integrates MERFISH, a fluorescence-based spatial transcriptomics method, to detect CRISPR-guided RNAs and gene transcripts. A key innovation was developing a signal amplification strategy, allowing scientists to observe small molecular changes with high precision.
In a proof-of-concept study, researchers measured the effects of 35 genes regulating immune cell response, finding strong alignment with previous Perturb-seq data. They also discovered that cell density influences gene perturbations, demonstrating how non-perturbed cells can be affected by nearby genetic modifications.
The research team aims to refine Perturb-FISH by making it more accessible and capable of analyzing more genes per experiment. The goal is to simplify the technique, requiring only a single imaging instrument, and expand the technology’s reach to more scientific fields. This breakthrough in spatial genetic research provides scientists with new tools for studying gene interactions in tissue development, disease progression, and therapeutic interventions.