Key Points
- AI discovers 70,500 previously unknown RNA viruses using metagenomics.
- The new viruses come from extreme environments and show unique biodiversity.
- The study uses AI models like ESMFold and LucaProt to identify viral RdRps.
- Further research is needed to determine the hosts of these newly discovered viruses.
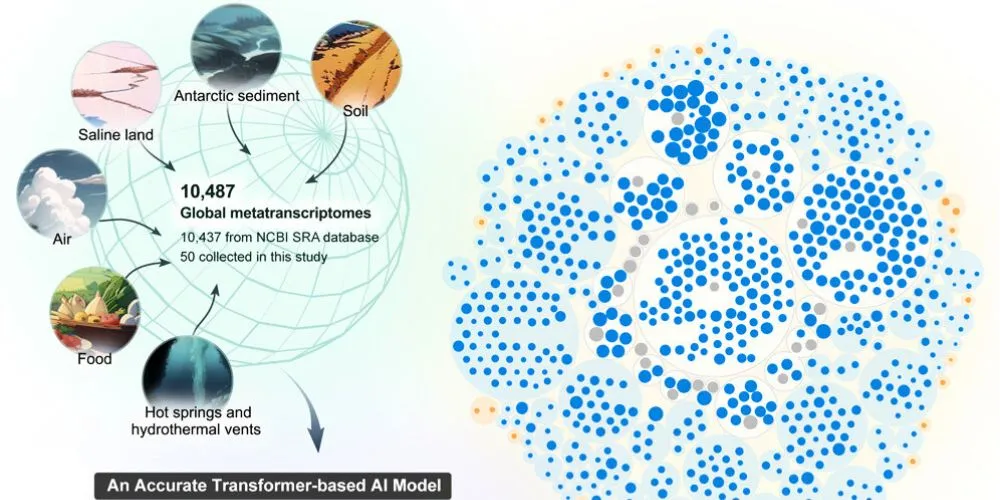
Researchers have used artificial intelligence (AI) to uncover more than 70,500 previously unknown RNA viruses, many unlike any species currently identified. This breakthrough highlights the potential of AI to explore the “dark matter” of the RNA virus world, significantly expanding the known virosphere. The discovery was made possible through metagenomics, where scientists sample all genomes in environmental samples, bypassing the need to culture individual viruses.
RNA viruses are abundant microorganisms that infect various hosts, including animals, plants, and even bacteria. However, only a few of these viruses have been identified and studied. Artem Babaian, a computational virologist at the University of Toronto, notes a “bottomless pit” of viruses left to be discovered. Identifying these viruses could be crucial in understanding the mysterious illnesses that some unknown viruses may cause.
The research, published in Cell, uses AI to delve deeper into virus discovery by analyzing predicted protein structures. The AI model incorporates ESMFold, a protein-prediction tool developed by Meta (formerly Facebook). A similar tool, AlphaFold, created by Google DeepMind, recently won the Nobel Prize in Chemistry. By using these advanced AI tools, researchers can better identify RNA viruses that traditional methods may have missed due to their rapid evolution.
In 2022, Babaian and his team discovered nearly 132,000 new RNA viruses through genomic sequencing data. However, traditional methods often fail to identify many RNA viruses, especially if their genetic sequences differ significantly from known viruses.
Evolutionary biologist Shi Mang from Sun Yat-sen University in China and his team developed the LucaProt model to overcome this. This AI model uses the transformer architecture behind ChatGPT, along with protein-prediction data, to recognize viral RNA-dependent RNA polymerases (RdRps), a key enzyme in RNA virus replication.
Using LucaProt, the researchers identified roughly 160,000 RNA viruses, including some from extreme environments like hot springs and salt lakes. Nearly half of these viruses were previously unknown to science. The study also revealed RNA viruses with unique biodiversity, offering insights into evolutionary branches that had not been explored.
Although the hosts of these newly discovered viruses remain unidentified, the research opens new avenues for understanding virus origins, evolution, and environmental roles. Shi is currently developing a model to predict which organisms these viruses infect, potentially offering further insights into their ecological impact.