Key Points:
- LMU researchers develop pMINFLUX multiplexing, enabling simultaneous tracking of rapid dynamic processes involving multiple molecules.
- The method leverages MINFLUX super-resolution microscopy, achieving localizations with one-nanometer precision.
- pMINFLUX registers time differences between dye excitation and fluorescence emission, providing insights into fluorescence lifetimes.
- The method enables accurate DNA strand tracking and measuring distances between biomolecule binding sites.
Researchers at Ludwig Maximilian University of Munich (LMU) have devised an innovative method to monitor rapid dynamic processes involving multiple molecules at the molecular scale simultaneously, marking a significant advancement in super-resolution microscopy techniques.
Within biological systems, intricate processes unfold through the interactions of various biomolecules, such as proteins and DNA. These dynamic events occur on a scale as small as a few nanometers, posing a challenge for traditional fluorescence microscopy techniques, which are limited by diffraction to resolutions of around 200 nanometers.
To overcome this resolution barrier, super-resolution microscopy methods have relied on the principle of fluorescence blinking, which temporally separates the fluorescence emission from different molecules, enabling localization below the classical resolution limit. However, this approach hampers the simultaneous tracking of multiple molecules, limiting the temporal resolution for dynamic processes.
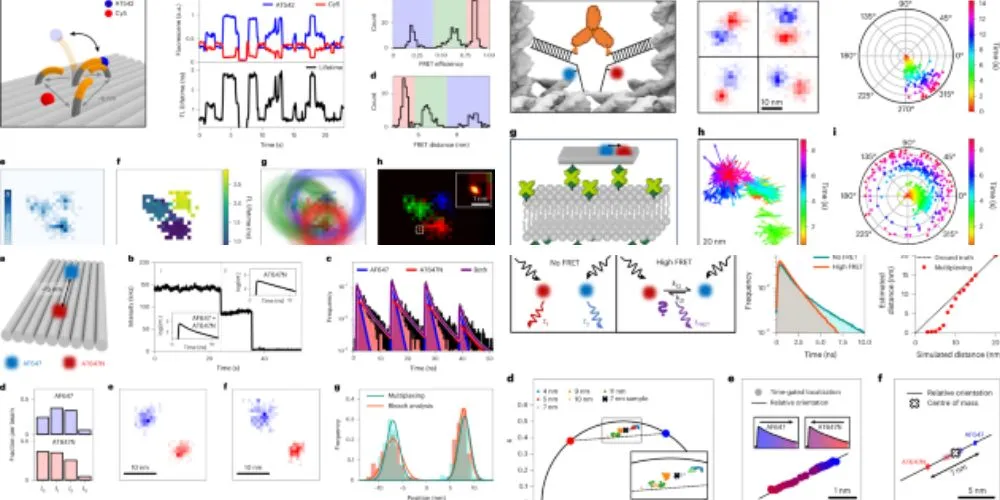
Led by LMU chemist Professor Philip Tinnefeld, in collaboration with Professor Fernando Stefani from Buenos Aires, the research team has developed pMINFLUX multiplexing, an elegant solution to this challenge. Their method, recently published in the journal Nature Photonics, leverages the MINFLUX super-resolution microscopy technique to achieve localizations with precision down to just one nanometer.
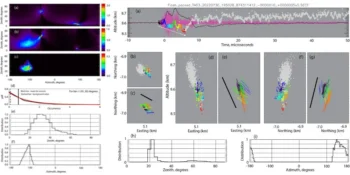
Unlike conventional MINFLUX, pMINFLUX incorporates a novel approach that registers the time difference between dye excitation and subsequent fluorescence emission with sub-nanosecond resolution. This enables precise localization of molecules and provides insights into their fluorescence lifetimes, a fundamental property affecting the duration of fluorescence emission.
By exploiting differences in fluorescence lifetimes among different dyes, the researchers devised a method to assign fluorescent photons to specific molecules without relying on blinking-induced temporal separation. By adapting to the localization algorithm and including a multiexponential fit model, they achieved simultaneous tracking of multiple dyes with nanometer precision, facilitating the investigation of rapid dynamic processes involving multiple biomolecules.
The researchers demonstrated the efficacy of pMINFLUX multiplexing by accurately tracking DNA strands on nanostructures, separating translational and rotational movements, and measuring distances between antibody-binding sites. Professor Tinnefeld anticipates that this breakthrough method will offer new insights into protein interactions and other biological phenomena.