Key Points:
- Researchers unveil a novel platform for identifying proteins capable of controlling protein stability, offering innovative therapeutic avenues.
- The study identifies over 600 new effector proteins with potential applications in targeted protein degradation and stabilization.
- Specific effectors like KLHL40, FBXL12, and FBXL15 hold promise for treating skeletal muscle disorders and chronic myeloid leukemia.
- The synthetic screening platform addresses the “protein pair problem” by enabling rapid and large-scale testing of effector-target protein interactions.
Researchers at Sinai Health and the University of Toronto have pioneered a groundbreaking platform to identify proteins capable of controlling the stability of other proteins, presenting a new avenue for disease treatment. This innovative approach seeks to leverage cellular processes for therapeutic purposes by pinpointing “effector” proteins that influence the stability of target proteins via induced proximity.
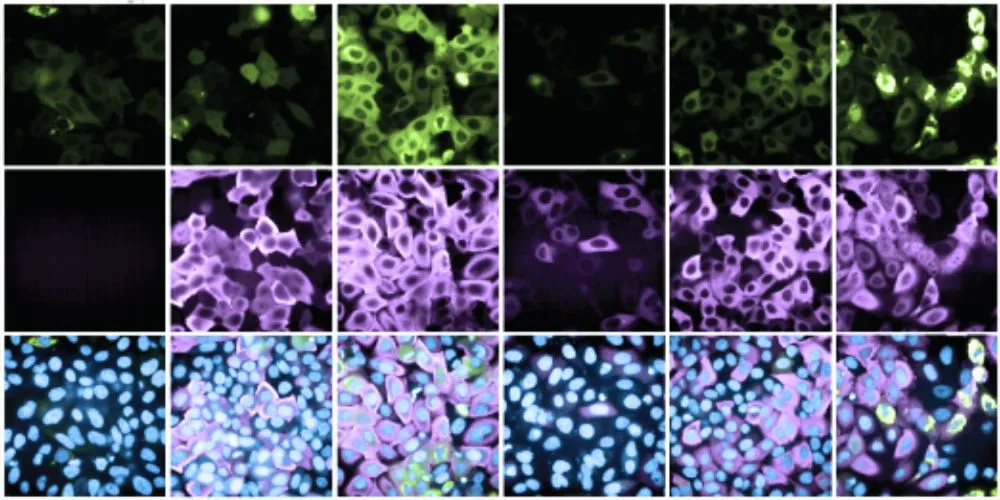
In a milestone achievement, the researchers devised a method to scrutinize the entire human proteome for potential effector proteins, uncovering over 600 new candidates within 14,000 genes. Remarkably, more than 200 of these newly identified effectors demonstrated proficiency in efficiently degrading target proteins, while approximately 400 exhibited the ability to stabilize target proteins, thereby enhancing their abundance.
Published in the journal Nature, this study marks the first comprehensive exploration of effector proteins on such a scale, offering a treasure trove of new candidates for drug development. The team’s synthetic platform enables unbiased, proteome-wide induced-proximity screens, facilitating the ongoing expansion of the effector protein library.
Traditionally, effector proteins employed for targeted protein degradation and stabilization include E3 ubiquitin ligases (E3s) and deubiquitinases (DUBs). However, the study revealed a surprising diversity among E3s in their efficacy in degrading target proteins. Notably, the discovery of “angry E3s” underscores their consistent ability to degrade targets across different cellular contexts.
Furthermore, the research team identified E2 conjugating enzymes as potent effectors for targeted protein degradation, challenging the conventional notion that only E3s are suitable. These findings highlight the untapped potential of E2 enzymes, previously overlooked in drug targeting due to perceived challenges.
The implications of this research extend to various therapeutic interventions. For instance, KLHL40, one of the identified effectors, holds promise for targeted protein stabilization in treating skeletal muscle disorders. Similarly, effector proteins FBXL12 and FBXL15 offer potential applications in targeted protein degradation for managing chronic myeloid leukemia.
The study’s synthetic screening platform addresses the “protein pair problem,” enabling rapid and large-scale testing of effector-target protein interactions. By leveraging an unbiased induced-proximity approach, researchers aim to overcome challenges associated with identifying suitable effectors for specific target proteins, thereby advancing the field of targeted therapies.