Key points
- Researchers identified an ancient protein, SCORE, that recognizes cold-shock proteins found in most bacteria, fungi, and insects.
- SCORE can be engineered to recognize and defend against a wide range of pathogens.
- This discovery offers a sustainable approach to enhancing disease resistance in crops.
- The protein’s variations exist across many flowering plant lineages, suggesting broad applicability.
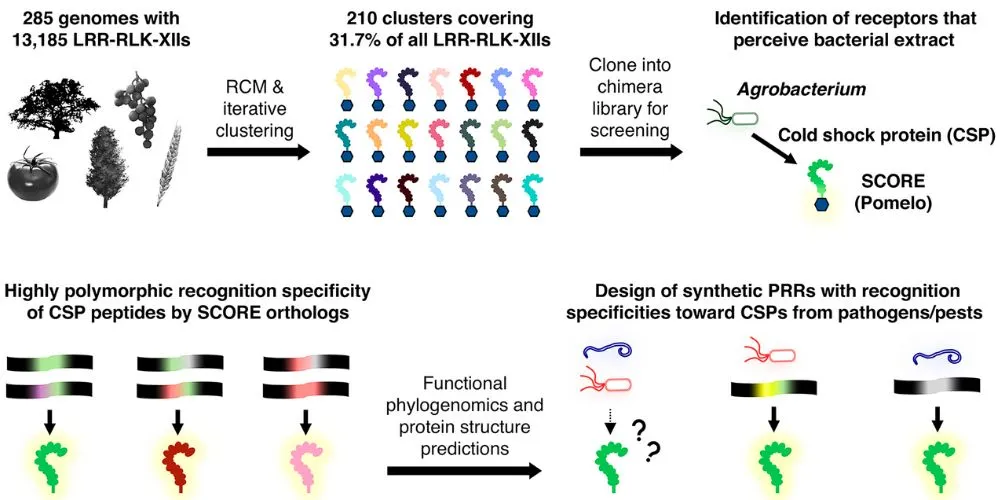
Scientists at the RIKEN Center for Sustainable Resource Science (CSRS) in Japan have unearthed a powerful tool in the fight against plant diseases. They’ve discovered an ancient protein, dubbed “SCORE” (Selective Cold Shock Protein Receptor), which acts as a highly adaptable immune receptor in plants.
SCORE detects cold-shock proteins, ubiquitous molecules found in over 85% of known bacteria, as well as fungi and insects. This makes it a potentially broad-spectrum defense mechanism against a vast array of plant pathogens.
The groundbreaking research, published in Science, reveals SCORE’s remarkable versatility. By simply altering specific amino acid sequences within SCORE, researchers can predictably change which cold-shock proteins it recognizes, effectively tailoring the protein’s pathogen-fighting capabilities.
This means SCORE can be engineered to target specific bacterial, fungal, or insect threats affecting various crops. Currently, plants often succumb to pathogens, resulting in reduced size and yield. The discovery of SCORE offers a promising solution to this widespread agricultural problem.
The researchers identified SCORE in pomelo, a citrus plant, after analyzing over 1,300 receptors from 350 plant species. They determined that SCORE recognizes a short 15-amino-acid section of cold-shock proteins (csp15). Remarkably, variations of SCORE exist across numerous plant lineages, tracing its origin back to the last common ancestor of flowering plants.
This indicates a long evolutionary history and potential for widespread application in various crops. Furthermore, the team found that specific amino acid locations within both SCORE and csp15 determine recognition, allowing for predictable engineering of new variants.
The implications for agriculture are profound. The team successfully engineered a version of SCORE that effectively targets pathogens like Ralstonia, Erwinia, and Xanthomonas, which naturally evade the original pomelo SCORE. This opens exciting avenues for creating disease-resistant crops through genetic engineering, potentially revolutionizing agricultural practices and enhancing global food security.
The researchers are now focusing on introducing engineered SCORE variants into economically important crop species to achieve broad-spectrum pathogen resistance, offering a more sustainable approach to disease management compared to traditional chemical methods.